The next time someone catches you playing a video game and asks what you are doing, you can now say that you are helping to save lives. Sure, that explanation may not save your job or convince your significant other that you are doing good via your PC, but for some it is actually true.
Scientists from the University of Washington recently decided to try a different approach to help them understand the detailed molecular structure of a retroviral proteases enzyme known as M-PMV that is instrumental in spreading AIDS, as well as other diseases. For over a decade now researchers have looked for the exact chemical key that would help recreate the enzyme, but with no luck.
For each enzyme being studied, there are millions of possible combinations to how the bonds between atoms form. The correct chemical key is the one that uses the most-efficient configuration with the lowest amount of energy. Once that has been unlocked, it can lead researchers to a better understanding of the enzyme, and a means to attack it.
There are computer programs that can help in tracking down the structure of proteins, but for more than ten years this particular simian-based enzyme has stumped scientists. Although computers have plenty of processing power to analyze the enzyme, they lack the spatial reasoning that is necessary to solving enzyme puzzles. That is where gamers come in.
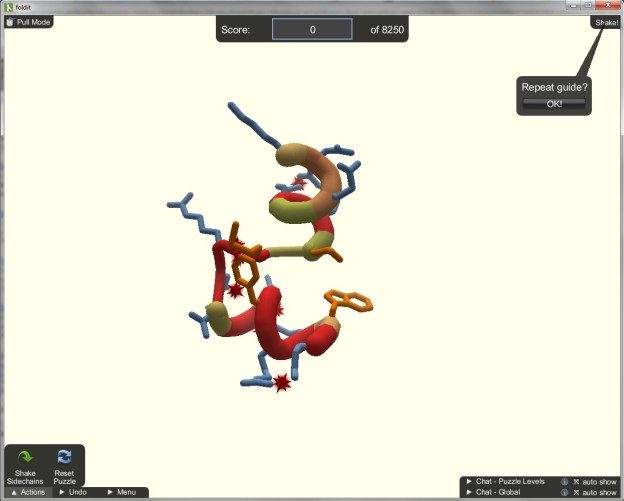
In what University of Washington researchers saw as a longshot at best, they sent a virtual model of the protein to fold.it, a program/website that asks gamers for help in solving complex protein puzzles that it displays like a game. In the case of this particular enzyme, what had stumped the scientific community for more than ten years was solved in ten days.
“This was really kind of a last-ditch effort. Can the Foldit players really solve it?” Firas Khatib, a biochemist at the University of Washington and the lead author on the recently published research paper told MSNBC. “They actually did it in less than 10 days.”
The name “Foldit” comes from the nature of the game, where users are asked to undertake one of the most difficult biological problems, folding proteins. The games themselves are open to anyone, and a background in biology is not necessary. Besides actually helping to solve specific protein puzzles, the users’ actions are also recorded and studied to help increase a computer’s ability to increase spatial reasoning through detailed algorithms created based on the games’ results.
Put simply (very, very simply), the game revolves around protein biosynthesis, the process in which cells make proteins. Scientists, specifically biologists, are all over this. Where it gets tricky is understanding how certain proteins became three-dimensional structures—or how the molecule is “folded.” The game itself is scored on three factors: How well you pack the protein, how efficiently you hide the hydrophobics and how you clear the clashes. If none of those phrases mean much to you, then don’t worry. The game is designed so anyone playing will see the protein folding as a game, complete with scores and leaderboards.
When a new protein is loaded up, gamers attempt to manipulate the structure in order to find its ideal form. With each correct move you earn points. When connected online, gamers are able to chat with other connected users in order to compare strategies in order to find the ideal form. In the case of M-PMV, gamers around the world built off of each others’ work to solve the puzzle.
Foldit originally debuted in beta back in 2008, and has since gone on to register close to 240,000 users.
Now that the M-PMV structure has been cracked, Seth Cooper, a University of Washington computer scientist and lead designer and developer of Foldit, is hoping that more scientists send them problems that fit within the Foldit format.
“The critical role of Foldit players in the solution of the M-PMV [retroviral protease] structure shows the power of online games to channel human intuition and three-dimensional pattern-matching skills to solve challenging scientific problems,” said the study, which was published by Nature Structural & Molecular Biology. “Although much attention has recently been given to the potential of crowdsourcing and game playing, this is the first instance that we are aware of in which online gamers solved a longstanding scientific problem.”


